
How it works
CB-Dock2 is an improved version of the protein-ligand blind docking tool that inherits the curvature-based cavity detection procedure and the AutoDock Vina-based molecular docking procedure in CB-Dock server (see CB-Dock for details). On this foundation, we future integrated a homologous template-based blind docking procedure, considering that the pocket information on the templates can provide valuable references for binding sites prediction in the presence of homologous templates. The complete workflow of CB-Dock2 is shown below. For the protein and ligand submitted by user, CB-Dock2 will retrieve from the protein-ligand complexes database stored on our server for template ligands with high topology similarity (FP2 ≥ 0.4) at the first time. If present, the similarity between the query protein and the proteins complexed with the selected template ligands will be calculated. The complexes with greater than 40% sequence identity (and pocket RMSD ≤ 4Å) in the template ligand binding site will be retained for the subsequent template-based cavity detection and molecular docking. The docking method utilized in this procedure is FitDock, an in-house developed method that fits initial conformation to the given template using a hierarchical multi-feature alignment approach, subsequently explores the possible conformations, and finally outputs refined docking poses.
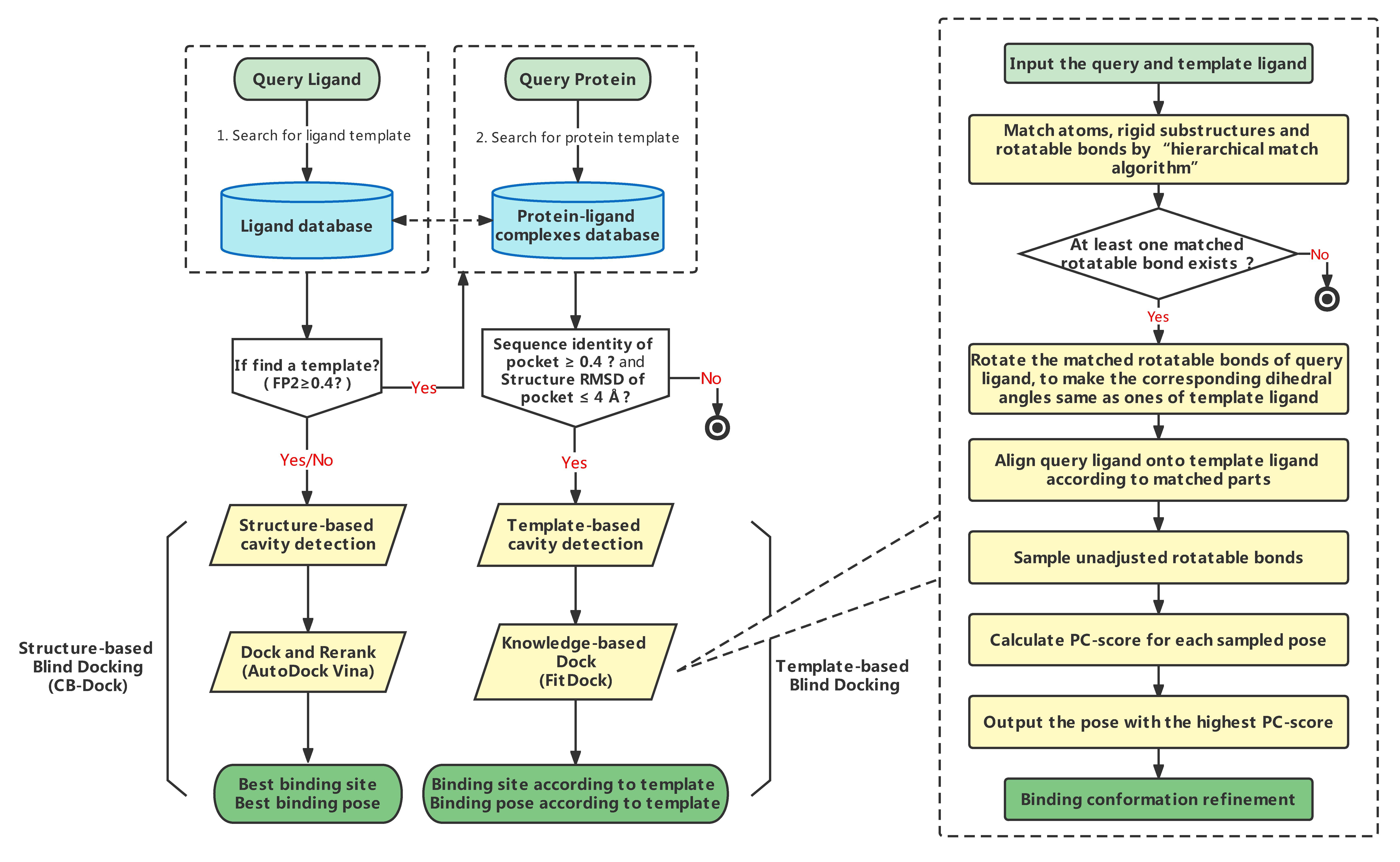
After submitting the required files, a perl script will process these files automatically as shown above. We employ the latest version (1.2.0) of AutoDock Vina for template-independent blind docking. The pipeline of template-based blind docking employs the BioLip2 database (version of 2025.04.23) as the template database.
Submit jobs
To meet the different demands of users, we designed two sets of task flow with different objectives, namely "Search Cavities" and "Auto Blind Docking". The former will perform cavity detection according to the protein and ligand submitted by the user, including structure-based cavity detection and template-based cavity detection (performed in the case of having homologous templates). The latter will automatically use the above two programs to perform cavity detection and then docking based on the detected pockets. Submission methods and notes are as follows:
- Query protein:
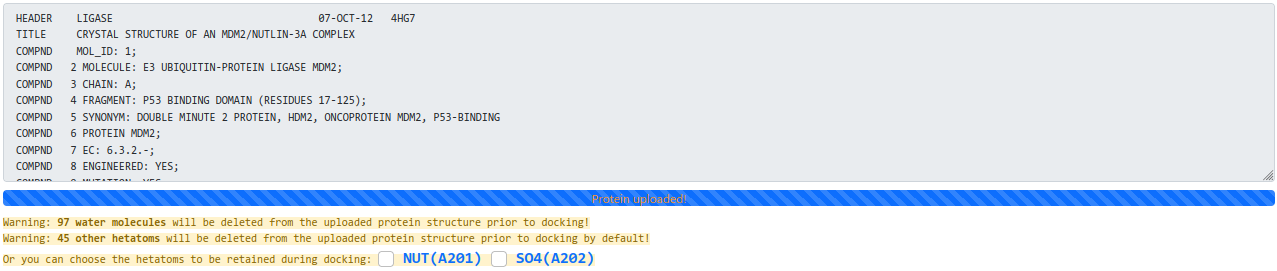
- The target protein is uploaded in PDB format [Example file]. We will remove all heteroatoms (including small molecules, ions, and water) of the protein before docking by default.
And you can pay attention to the warning information given by the protein submission zone (as shown below), to know if some hetatoms are present in your submitted protein. Furthermore, you can choose the hetatoms listed in the warning information to be retained during docking.
 Hint: If the protein is a biological assembly, you can split it into chains before docking (Split Chains).
Hint: If the protein is a biological assembly, you can split it into chains before docking (Split Chains). - Query ligand:
- The ligand can be uploaded in MOL2, MOL, SDF, or PDB format [Example file]. If the file format is MOL or SDF, we will convert it to PDB using Open Babel.
Alternatively, you can draw a ligand with the JSME Molecule Editor embedded in our webserver.
Hint: The ligand file size should be less than 15KB, because the accuracy and efficiency of AutoDock Vina will be reduced as the number of atoms and rotatable bonds increase.
- Number of cavities for detecting:
- The Number of cavities detected by the template-independent method, and the default value is 5. Users can click "More parameters" to customize this value. The program will perform docking in each cavity and rank the cavities according to the best Vina score.
Hint: The number of cavities should be less than 20, because greater numbers will increase the runtime and the false positive rate.
- Tempalte:
- This is an optional parameter, and users can choose to upload protein-ligand complex for template-based blinding docking.
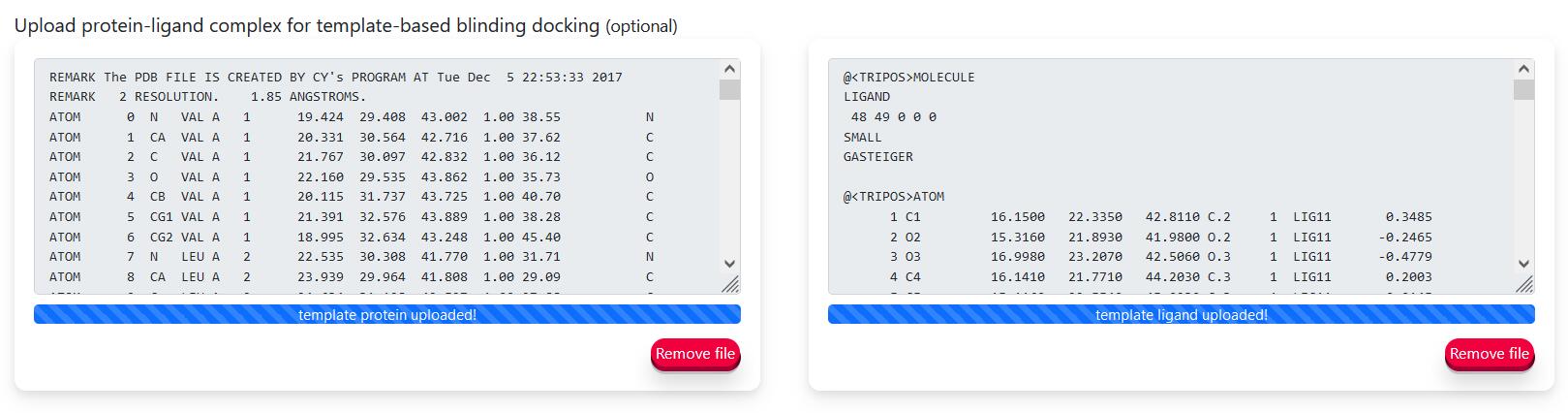 Hint: If the protein-ligand template is a biological assembly, you can split it into chains before uploading (Split Chains).
Hint: If the protein-ligand template is a biological assembly, you can split it into chains before uploading (Split Chains).
Users can submit one "Search Cavities" job or "Auto Blind Docking" job at a time. Once the task is completed, you can click on the "View Result" in the task list to jump to the result page, then you can view or download the results under the result page. Or you can delete the task output from our server by clicking the "Remove" button on the task list. Additional, We will send the result email if users enter the eamil address.
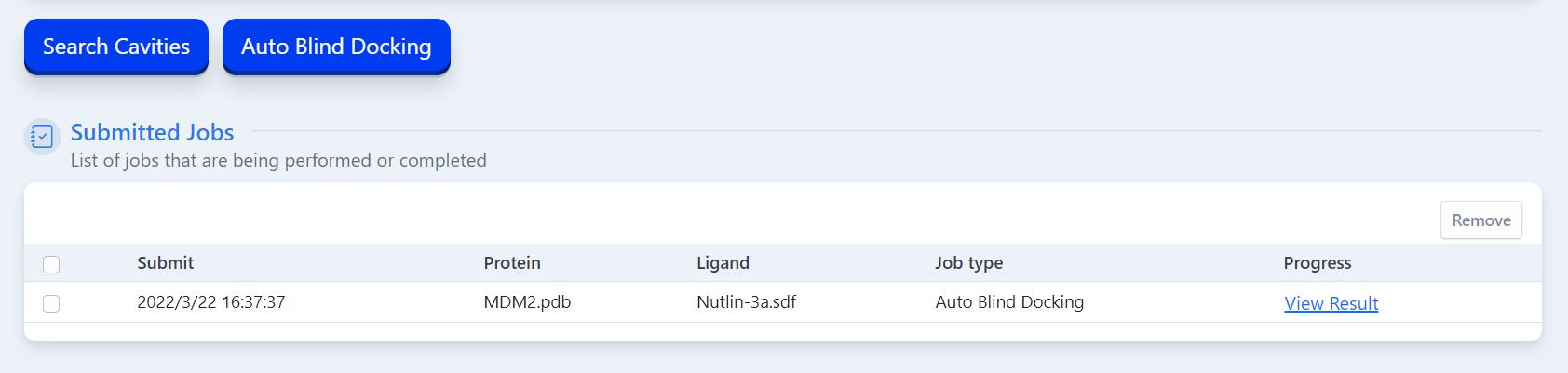
Tasks that are not manually deleted will be automatically deleted after one day, so please download and save your task output in time.
View results
For the two different tasks of "Search Cavities" and "Auto BlindDock", we provide plentiful visualization functions on the results page to help users preliminarily analyze the calculation results. Users can also download the data locally for further analysis.
- Results of Search Cavities:
-
The cavity detection result interface is divided into two parts according to the detection method: Structure-based cavity detection and template-based cavity detection (performed in the case of having homologous templates).
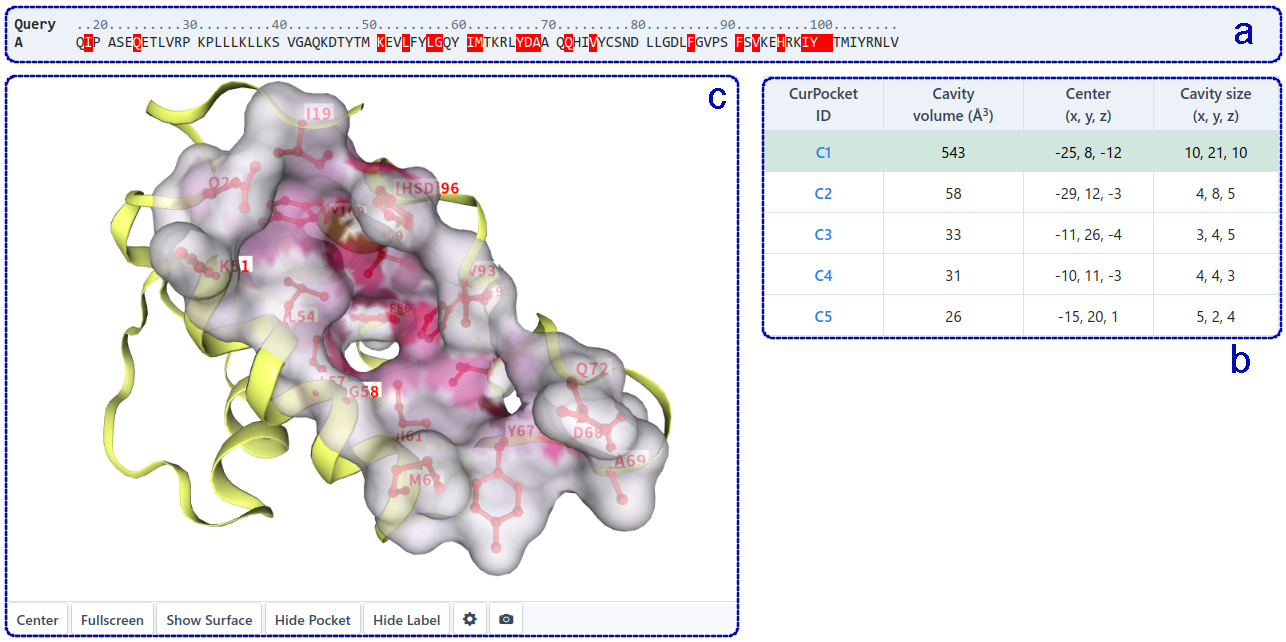
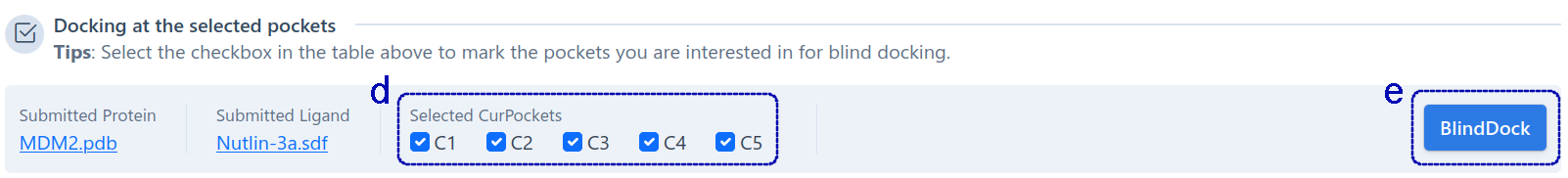
For structure-based cavity detection (as shown below), this section shows the whole sequence (a) of the query protein, and highlights the residues in each detected cavities (b). Users can click the residue in the sequence list to show it in 3D-view (c). The detected cavities is sorted by volume, and users can click on different cavity in the table to view the detailed structure information in 3D-view. In addition, users can check the cavities of interest (d) for molecular docking based on AutoDock Vina(e), so as to obtain the potential binding modes of query ligand in the target cavities.
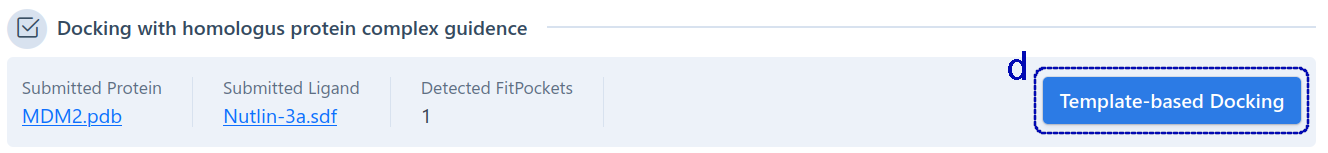
For template-based cavity detection (as shown below), this section shows the sequence alignment (a) of the query protein and the template protein, and highlights the pocket residues detected based on the template (b). Similarly, users can click the residue in the sequence list to show it in 3D-view (c). The detected pockets are first sorted according to FP2. If FP2 is the same, they are sorted by pocket identity. The pockets detected from different template, but with the same "FitPocket ID" in the table (b), have at least 50% pocket residues overlapped. In addition, our server automatically gives the consensus analysis between the pockets detected based on template and the pockets detected based on structure. Users can click the "Template-base Docking" (d) to view the binding modes of query ligand in the target pockets.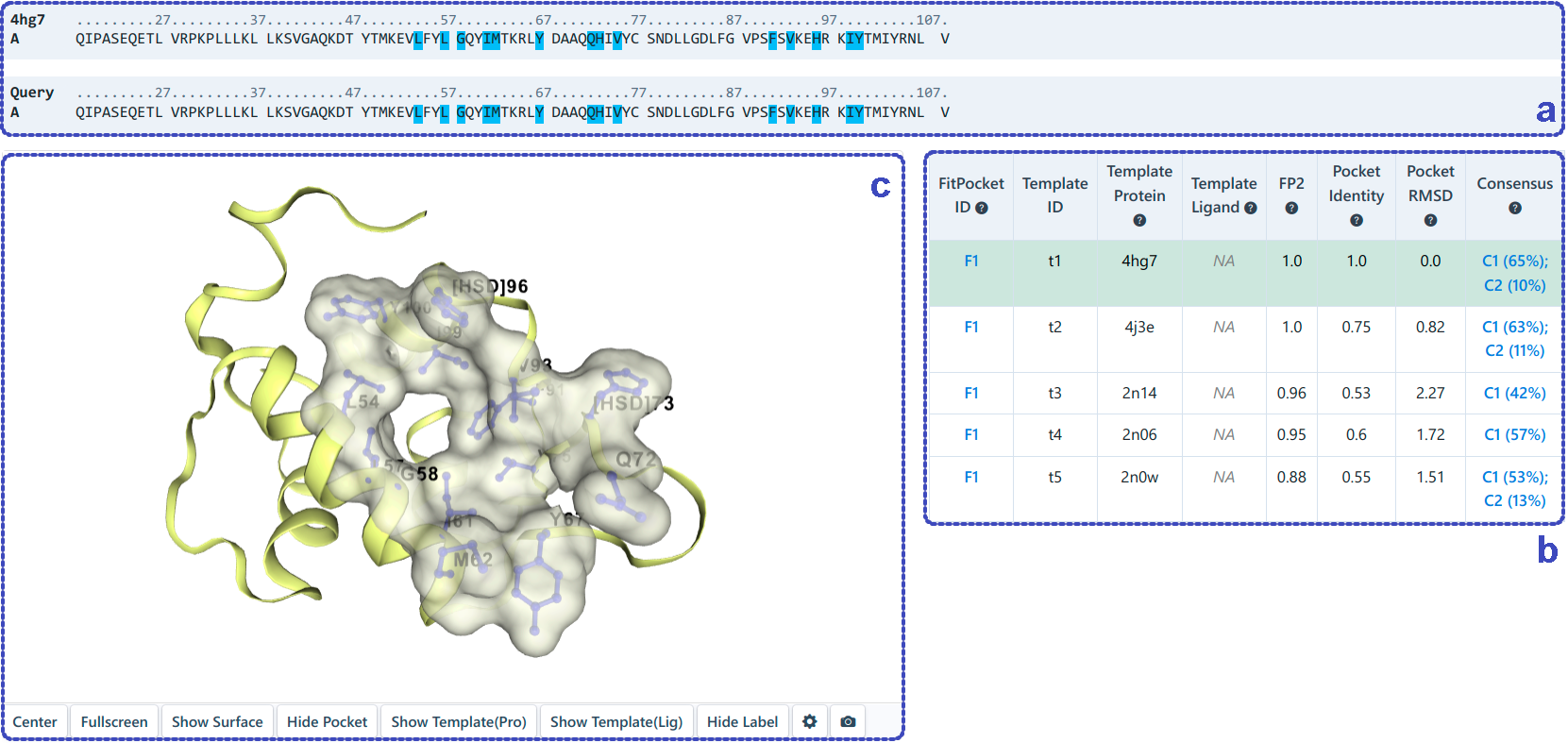
- Results of Auto BlindDock:
-
The result interface of auto blind docking gives the predicted results based on two different blind docking principles.
For structure-based blind docking (as shown below), the docking based on AutoDock Vina will be performed in each cavity detected. We rank the potential binding sites of the query ligand according to the Vina score (kcal/mol). Users can click the "View" button in the table (a) to view the contact residues (c) of the best binding mode in each pocket, combining the structural information in 3D-view (b).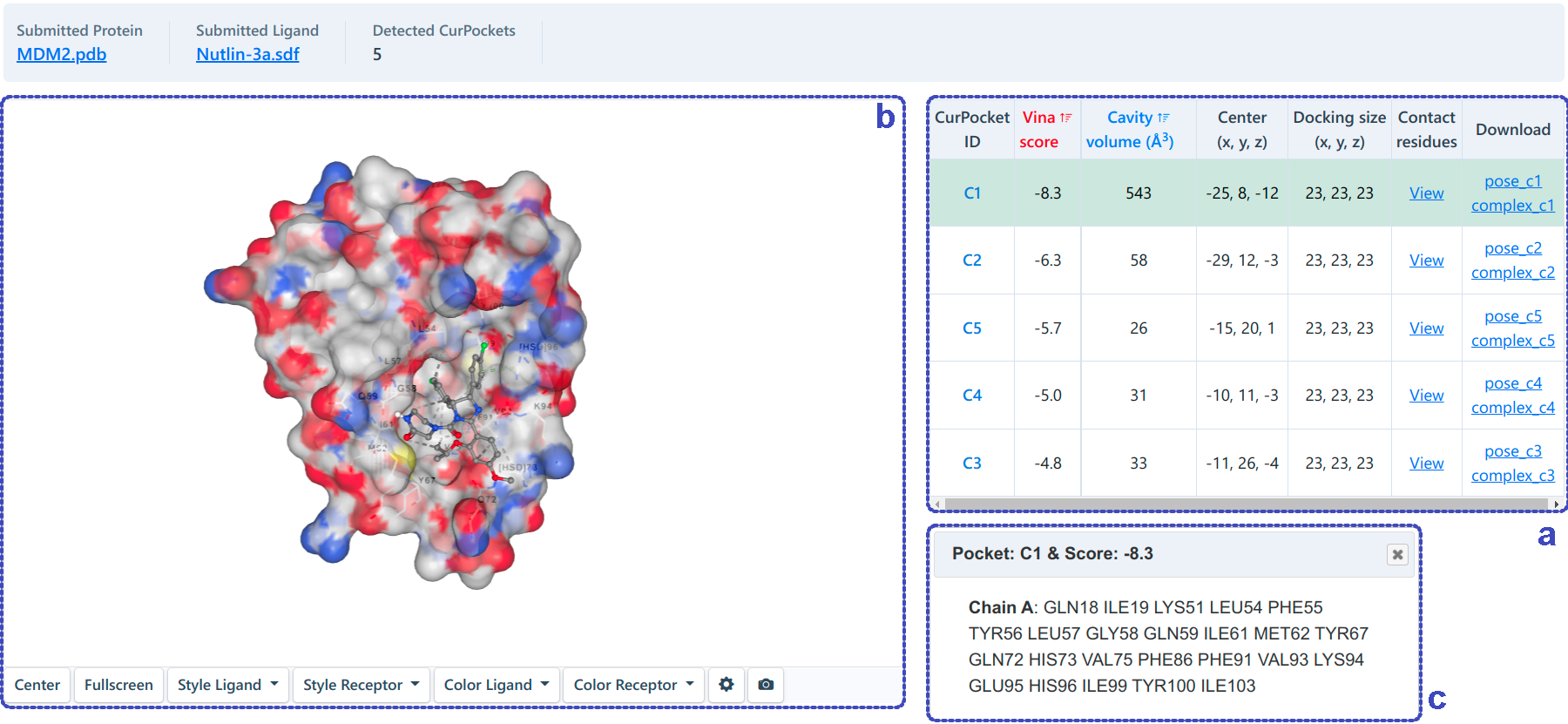
For template-based blind docking (as shown below), knowledge-based docking developed in house will be performed in each cavity detected. Similarly, we rank the potential binding sites of the query ligand according to the docking score (kcal/mol). The binding modes with the same "FitPocket ID" belong to different binding conformation in the same pocket. Users can determine the best binding conformation at a site according to the docking score, templates and the contact information. The tempaltes (c) and contact information (d) can be achieved from the table (a) and the 3D-view (b) by clicking the corresponding button.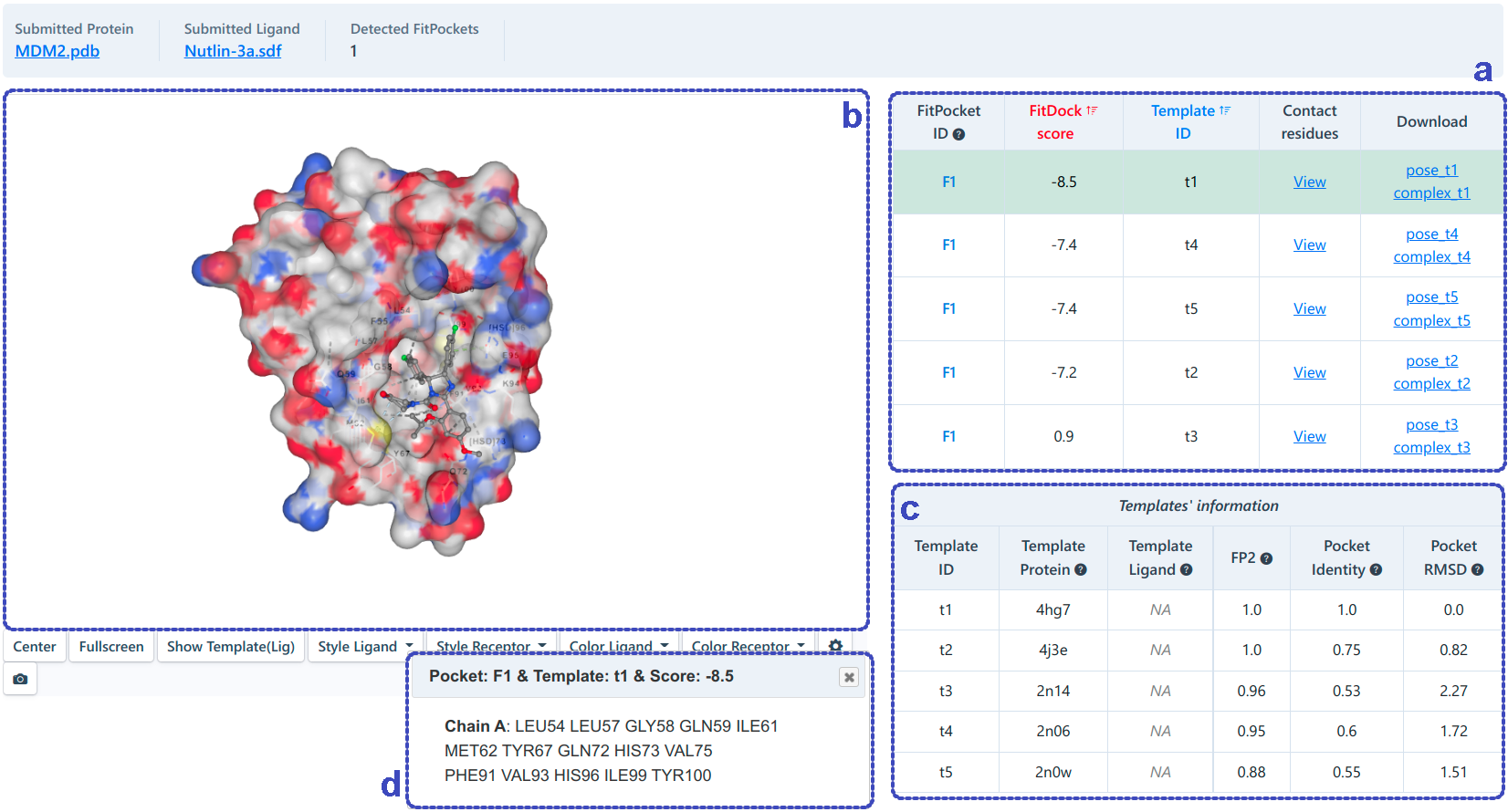
The meta-analysis integrated the results of the above two blind docking methods (as shown below). The cavities detected based on the above two methods will be clustered based on the overlap ratio of cavity residues (a, b). Since the structure-based and template-based blind docking methods may detect the same binding site and predict the binding poses of the query ligand based on AutoDock Vina and FitDock, respectively. In this situation, how to determine the best binding pose may be a concern for users. In case the templates are extracted, the RMSD between the best poses obtained by the two blind docking methods will be calculated, and if the RMSD is less than 2 Å, the pose with the lower score is chosen as the best prediction, otherwise the pose generated by template-based blind docking is selected as the best prediction. In the absence of templates, the pose obtained by structure-based blind docking is selected as the best prediction (a, b). Users can preview the integrated pockets and the best binding pose for each pocket via NGL-viewer (c).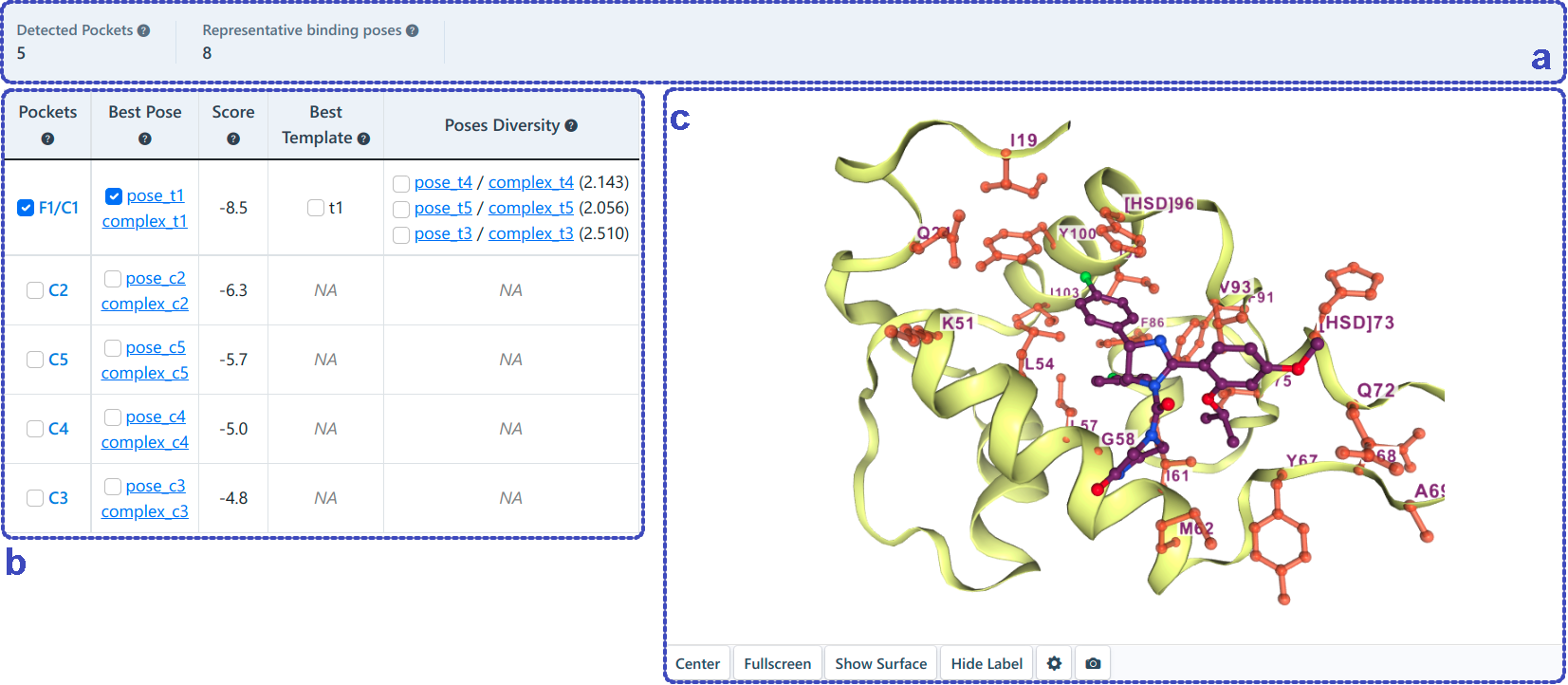
- Contact type in NGL-viewer:
-
Type Color Style Description Criterion Citations Hydrogen Bond 
Hydrogen-bond between strong donor and acceptor atoms. maxHbondDist: 3.5 Å
maxHbondSulfurDist: 4.1 Å
maxHbondAccAngle: 45 Å
maxHbondDonAngle: 45 Å
maxHbondAccPlaneAngle: 90
maxHbondDonPlaneAngle: 30
[1] Sulfur-containing H-bonds
[2] H-bondsWeak Hydrogen Bond 
Hydrogen-bond between a carbon donor atom and an acceptor, or a Pi group and a donor atom. maxHbondDist: 3.5 Å
maxHbondSulfurDist: 4.1 Å
maxHbondAccAngle: 45 Å
maxHbondDonAngle: 45 Å
maxHbondAccPlaneAngle: 90
maxHbondDonPlaneAngle: 30
[1] Sulfur-containing H-bonds
[2] H-bondsHydrophobic Interaction 
Interactions between alkyl groups, or a alkyl group and a Pi group. maxHydrophobicDist: 4 Å Halogen Bond 
Interactions with fluorine, chlorine, bromine or iodine atoms. maxHalogenBondDist: 4 Å
maxHalogenBondAngle: 30Ionic Interaction 
Interactions between pairs of oppositely charged groups. maxIonicDist: 5 Å Cation-Pi Interaction 
Interactions between a positively charged atom and the electrons of a delocalized Pi system. maxCationPiDist: 6 Å
maxCationPiOffset: 2 ÅPi-Pi Stacking 
Interactions between delocalized Pi systems. maxPiStackingDist: 5.5 Å
maxPiStackingOffset: 2 Å
maxPiStackingAngle: 30
Issues summary
-
Browser Compatibility
We tested CB-Dock2 using the latest versions of IE, Edge, Firefox, Chrome and Safari. Browser compatibility is shown in the table below. If you are using one of the before mentioned browsers and you have problems displaying the “Results” page, we recommend you to update your browser to the latest version. Some browsers may not support WebGL or do not support all WebGL features needed by NGL viewer like Opera. Please try another browser in the case you get a warning that WebGL is not supported or the example provided by us on the “Results” page is not visible.
OS Chrome(v96) Firefox(v88)
Firefox(v88) Edge(v96)
Edge(v96) Safari
Safari
Linux (ubuntu18) Windows10 MacOS -
Why can't I receive the result email ?
The mail may be identified as spam. You may need to check in the trash can and add our account to the whitelist.
-
Why does the url in the results email not show the results ?
Your submission may have expired as the server only saves the results for one day.
-
Whether CB-Dock2 currently supports proteins carrying metal ions for docking ?
CB-Dock2 is currently unable to perform docking calculations on metal ion-containing proteins, and the corresponding functions need to be improved. So at present, in the protein submission interface, the metal ion component in the protein cannot be checked.
Publications
Yang Liu, Xiaocong Yang, Jianhong Gan, Shuang Chen, Zhi-Xiong Xiao, Yang Cao . CB-Dock2: improved protein-ligand blind docking by integrating cavity detection, docking and homologous template fitting. Nucleic Acids Research, 2022.
Xiaocong Yang, Yang Liu, Jianhong Gan, Zhi-Xiong Xiao, Yang Cao . FitDock: protein-ligand docking by template fitting. Briefings In Bioinformatics, 2022.
Yang Liu, Maximilian Grimm, Wen-Tao Dai, Mu-Chun Hou, Zhi-Xiong Xiao, Yang Cao . CB-Dock: a web server for cavity detection-guided protein–ligand blind docking. Acta Pharmacologica Sinica, 2019.
Yang Cao , Lei Li. Improved protein-ligand binding affinity prediction by using a curvature dependent surface area model. Bioinformatics, 2014.
Privacy policy
Data and results of each user are not accessible to other users, but we cannot guarantee full privacy.
Data retention policy
Data and results will be stored on the server only for a limited amount of time. The content of the server will be periodically deleted.
Currently we delete data after one day, users may directly delete the data stored on our server, using the Remove button under Dock.
We reserve the right to adjust the data retention policy depending on the workload of our server.
Cookie notice
In order to provide the CB-Dock2 service, this server uses cookies. The cookies are used solely for the purpose to display the correct results for each user. If you want to use the CB-Dock2, you need to accept the session cookie, otherwise the service can not be used. We don't use any third party cookies.
License
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.